Forward problem: (Inversion sampling)#
There are many different definitions for what is commonly called “The forward problem”. We will define the forward problem as the following task:
Given a (1) model and (2) parameter values then produce a feasible set of observations
Definition of a model#
What do we mean when we say “a model”? A model—in the most abstract terminology—is a collection of probability distributions \(\mathcal{P}\) over a sample space \(\mathcal{G}\) of potential measurements. In other words, we can define a model as the following two objects
where the input for \(F_{\theta}\), the cumulative density function (cdf), is all possible points in the sample space \(\mathcal{G}\). That is, we assume that \(F_{\theta}\) is a cdf that can input points like \((x_{1},x_{2}, \cdots, x_{k})\). Note that every point \(\theta \in \Theta\) will specify exactly one assignment of probabilities to all points in the sample space.
This definition of a model is abstract, but encompasses almost all feasible types of models that we can specify.
Example
Suppose we work for a public health office and are asked to begin modeling the incidence of influenza over during the typical 32-week influenza season. We assume that, for the person requesting this model, it is sufficient to provide a probability density over the potential number of weekly lab-confirmed cases of influenza in the public health office’s jurisdiction. The number of cases that we could observe (could measure) in one week starts at 0 (no cases this week) and end at the total number of individuals living in the jurisdiction (we will call this value \(N\)).
Then the sample space is \(\mathcal{G} = \{ (x_{1},x_{2}, \cdots, x_{32}) \;| \; x_{k} \in [0,N] \}\)
Further, we will assume that the number of cases each week is drawn from a Poisson distribution with parameter \(\lambda\). That is, for week \(k\), we assume \(x_{k} \sim \text{Poisson}(\lambda_{k})\). If we further assume that the number of cases in week \(k\) is statistically independent from cases in week \(l\) then the probability of measuring less than \(x_{k}\) cases in week \(k\) and less than \(x_{l}\) cases in week \(l\) equals
where \((\lambda_{k}, \lambda_{l}) \in \mathbb{R}^{+} \times \mathbb{R}^{+}\)
If we can write down our collection of probabilities for two points then we can write this collection for \(32\) points
We can generate a dataset—a tuple of possible measurements—from the above model if we are given a set of 32 parameter values and a method for drawing values from the Poisson distribution.
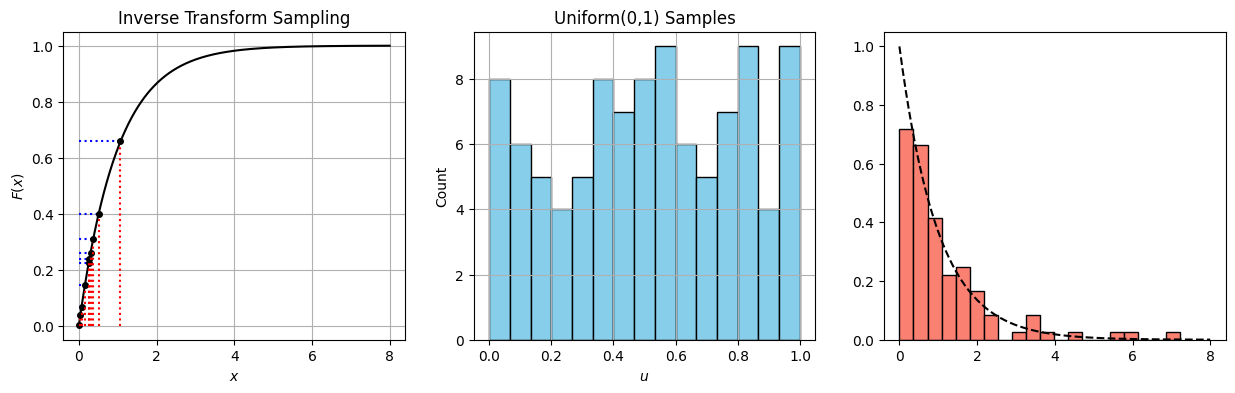
Inverse transform sampling#
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import expon
# --- Sampling ---
n_samples = 100
u_samples = np.random.uniform(0, 1, n_samples)
x_samples = -np.log(1 - u_samples)/1 # Inverse CDF of Exp(1)
# For plotting the CDF
x_grid = np.linspace(0, 8, 500)
cdf_vals = expon.cdf(x_grid)
# For highlighting transformation (only show a few)
u_highlight = np.sort(u_samples[:10])
x_highlight = -np.log(1 - u_highlight)
# --- PLOT ---
fig, axs = plt.subplots(1, 3, figsize=(15, 4))
# Plot 1: CDF with inverse transform lines
axs[0].plot(x_grid, cdf_vals, label='CDF of Exp(1)', color='black')
for u, x in zip(u_highlight, x_highlight):
axs[0].hlines(u, 0, x, color='blue', linestyle='dotted')
axs[0].vlines(x, 0, u, color='red', linestyle='dotted')
axs[0].plot(x, u, 'ko', markersize=4)
axs[0].set_title("Inverse Transform Sampling")
axs[0].set_xlabel("$x$")
axs[0].set_ylabel("$F(x)$")
axs[0].grid(True)
# Plot 2: Histogram of uniform(0,1) samples
axs[1].hist(u_samples, bins=15, color="skyblue", edgecolor="black")
axs[1].set_title("Uniform(0,1) Samples")
axs[1].set_xlabel("$u$")
axs[1].set_ylabel("Count")
axs[1].grid(True)
# Plot 3: Histogram of exponential samples
axs[2].hist(x_samples, bins=20, color="salmon", edgecolor="black", density=True, label="Samples")
axs[2].plot(x_grid, expon.pdf(x_grid), 'k--', label='True PDF') # overlay
[<matplotlib.lines.Line2D at 0x7f0225215410>]
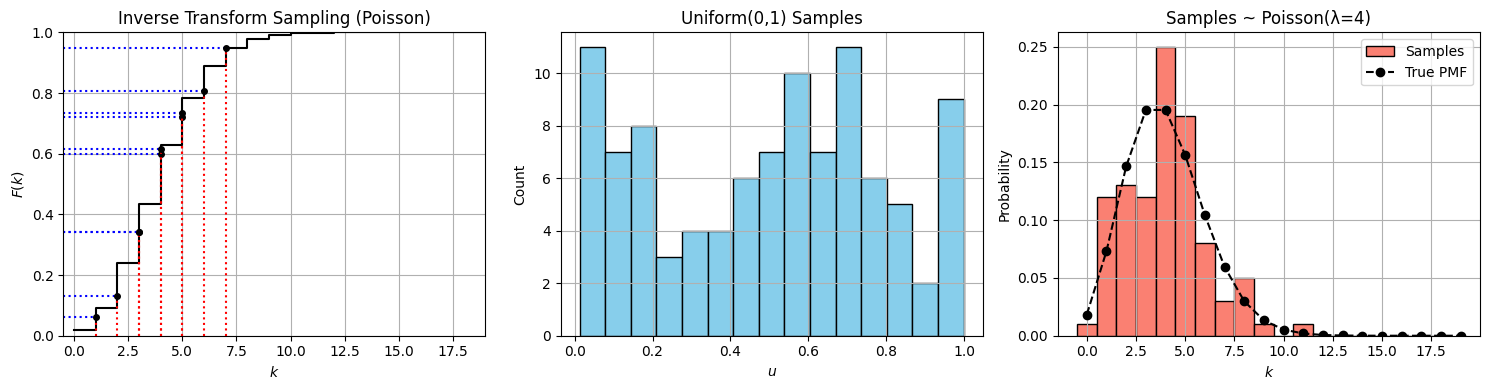
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import poisson
# Parameters
lam = 4 # Poisson mean
n_samples = 100
# Step 1: Generate uniform samples
u_samples = np.random.uniform(0, 1, n_samples)
# Step 2: Inverse transform sampling for Poisson
def poisson_inverse_transform(u, lam):
"""Perform inverse CDF sampling for Poisson(λ)."""
k = 0
cdf = poisson.pmf(k, lam)
while u > cdf:
k += 1
cdf += poisson.pmf(k, lam)
return k
x_samples = np.array([poisson_inverse_transform(u, lam) for u in u_samples])
# Step 3: CDF grid
k_vals = np.arange(0, 20)
cdf_vals = poisson.cdf(k_vals, lam)
# Use a few points for illustrating transformation
u_highlight = np.sort(u_samples[:10])
x_highlight = [poisson_inverse_transform(u, lam) for u in u_highlight]
# --- Plotting ---
fig, axs = plt.subplots(1, 3, figsize=(15, 4))
# Plot 1: CDF and mapping lines
axs[0].step(k_vals, cdf_vals, where='post', color='black', label='Poisson CDF (λ=4)')
for u, x in zip(u_highlight, x_highlight):
axs[0].hlines(u, -1, x, color='blue', linestyle='dotted')
axs[0].vlines(x, 0, u, color='red', linestyle='dotted')
axs[0].plot(x, u, 'ko', markersize=4)
axs[0].set_xlim(-0.5, max(k_vals))
axs[0].set_ylim(0, 1)
axs[0].set_xlabel('$k$')
axs[0].set_ylabel('$F(k)$')
axs[0].set_title("Inverse Transform Sampling (Poisson)")
axs[0].grid(True)
# Plot 2: Histogram of uniform samples
axs[1].hist(u_samples, bins=15, color="skyblue", edgecolor="black")
axs[1].set_title("Uniform(0,1) Samples")
axs[1].set_xlabel("$u$")
axs[1].set_ylabel("Count")
axs[1].grid(True)
# Plot 3: Histogram of Poisson samples
axs[2].hist(x_samples, bins=np.arange(0, 20) - 0.5, color="salmon", edgecolor="black", density=True, label="Samples")
axs[2].plot(k_vals, poisson.pmf(k_vals, lam), 'ko--', label='True PMF')
axs[2].set_title(f"Samples ~ Poisson(λ={lam})")
axs[2].set_xlabel("$k$")
axs[2].set_ylabel("Probability")
axs[2].legend()
axs[2].grid(True)
plt.tight_layout()
plt.show()
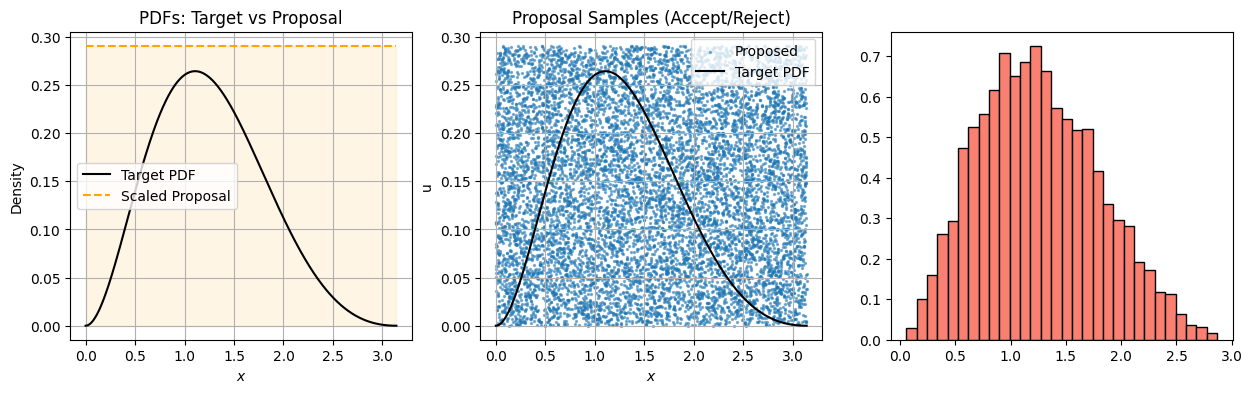
Accept-Reject Sampling: Derivation Using Conditional Densities#
We want to simulate from a target distribution ( f(x) ), but we only have access to samples from a proposal distribution ( g(x) ), where:
We define a uniform random variable ( U \sim \text{Uniform}(0, 1) ) and accept the sample ( x \sim g(x) ) if:
Joint Density Over ((x, u))#
We define the joint distribution over ( x ) and ( u ):
The pair ((x, u)) is accepted if:
Step 1: Joint Probability of Acceptance and ( X < x )#
We compute the joint probability of ( X < x ) and the sample being accepted:
Step 2: Marginal Probability of Acceptance#
We integrate over the full support:
Step 3: Conditional Distribution of ( X \mid \text{Accept} )#
Using the definition of conditional probability:
Therefore, the accepted samples ( x ) follow the desired distribution ( f(x) )!
import numpy as np
import matplotlib.pyplot as plt
# --- Target and Proposal ---
def target_pdf(x):
return np.exp(-x) * (np.sin(x)**2) # Not normalized
def proposal_pdf(x):
return np.ones_like(x) # Uniform on [0, π]
# --- Settings ---
N = 10000
x_min, x_max = 0, np.pi
x_grid = np.linspace(x_min, x_max, 500)
# Normalize target to get max for rejection
unnormalized = target_pdf(x_grid)
M = 1.1 * np.max(unnormalized) # Safety buffer
print(f"Using M = {M:.3f} for rejection bound")
# --- Rejection Sampling ---
proposal_samples = np.random.uniform(x_min, x_max, N)
proposal_heights = np.random.uniform(0, M, N)
target_vals = target_pdf(proposal_samples)
accepted = proposal_heights < target_vals
accepted_samples = proposal_samples[accepted]
# --- Plotting ---
fig, axs = plt.subplots(1, 3, figsize=(15, 4))
# Panel 1: Proposal and Target PDFs
axs[0].plot(x_grid, target_pdf(x_grid), label="Target PDF", color='black')
axs[0].plot(x_grid, M * proposal_pdf(x_grid), label="Scaled Proposal", color='orange', linestyle='--')
axs[0].fill_between(x_grid, 0, proposal_pdf(x_grid)*M, color='orange', alpha=0.1)
axs[0].set_title("PDFs: Target vs Proposal")
axs[0].set_xlabel("$x$")
axs[0].set_ylabel("Density")
axs[0].legend()
axs[0].grid(True)
# Panel 2: Proposal samples with rejection
axs[1].scatter(proposal_samples, proposal_heights, s=2, alpha=0.5, label="Proposed")
axs[1].plot(x_grid, target_pdf(x_grid), color='black', label='Target PDF')
axs[1].set_title("Proposal Samples (Accept/Reject)")
axs[1].set_xlabel("$x$")
axs[1].set_ylabel("u")
axs[1].legend()
axs[1].grid(True)
# Panel 3: Accepted Samples
axs[2].hist(accepted_samples, bins=30, density=True, color='salmon', edgecolor='black', label="Accepted Samples")
axs[2].plot(x_grid, target_pdf(x_grid) / np.trapz(target_pdf(x_grid), x_grid), 'k--', label="Target PDF (normalized)")
axs[2].set_title("Accepted Samples ~ Target")
axs[2].set_xlabel("$x$")
axs[2].set_ylabel("Density")
axs[2].legend()
axs[2].grid(True)
plt.tight_layout()
plt.show()
Using M = 0.291 for rejection bound
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[3], line 53
51 # Panel 3: Accepted Samples
52 axs[2].hist(accepted_samples, bins=30, density=True, color='salmon', edgecolor='black', label="Accepted Samples")
---> 53 axs[2].plot(x_grid, target_pdf(x_grid) / np.trapz(target_pdf(x_grid), x_grid), 'k--', label="Target PDF (normalized)")
54 axs[2].set_title("Accepted Samples ~ Target")
55 axs[2].set_xlabel("$x$")
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/numpy/__init__.py:792, in __getattr__(attr)
789 import numpy.char as char
790 return char.chararray
--> 792 raise AttributeError(f"module {__name__!r} has no attribute {attr!r}")
AttributeError: module 'numpy' has no attribute 'trapz'
Homework#
[Coding] Code up the Accept/Reject algorithm to sample from the following target distribution \(f_{X}(x) = x^{2}e^{-x}\). We will need to use a proposal distribution \(g(x)\) that is greater than all values for \(f_{X}(x)\). Lets choose as proposal distribution \(g(x) = \lambda e^{-\lambda x}\) with a \(\lambda\) value equal to one.
Present your code
Present a histogram of samples from g
Present a histogram of samples from f (ie accepted samples)
In the above example, the distribution \(f_{X}(x)\) is “unnormalized” or the integral over all possible values does not sum to one. This suggests that we can use the Accept/Reject algorithm if we know the unnormalized form of our probability density \(h_{X}(x) = d f_{X}(x)\) for some constant d. Please show using the steps we used previously, that \(P(Y|Accept) = f_{X}(x)\) if
\(Y \sim g(x)\)
\(U \sim \text{Unif}(0,1)\)
Accept a point from y if \(U < \frac{d f_{X}(x)}{c g(x)}\)
